Research Highlights
Automatic Segmentation of Organelles in Electron Microscopy Image Stacks: A New Workflow
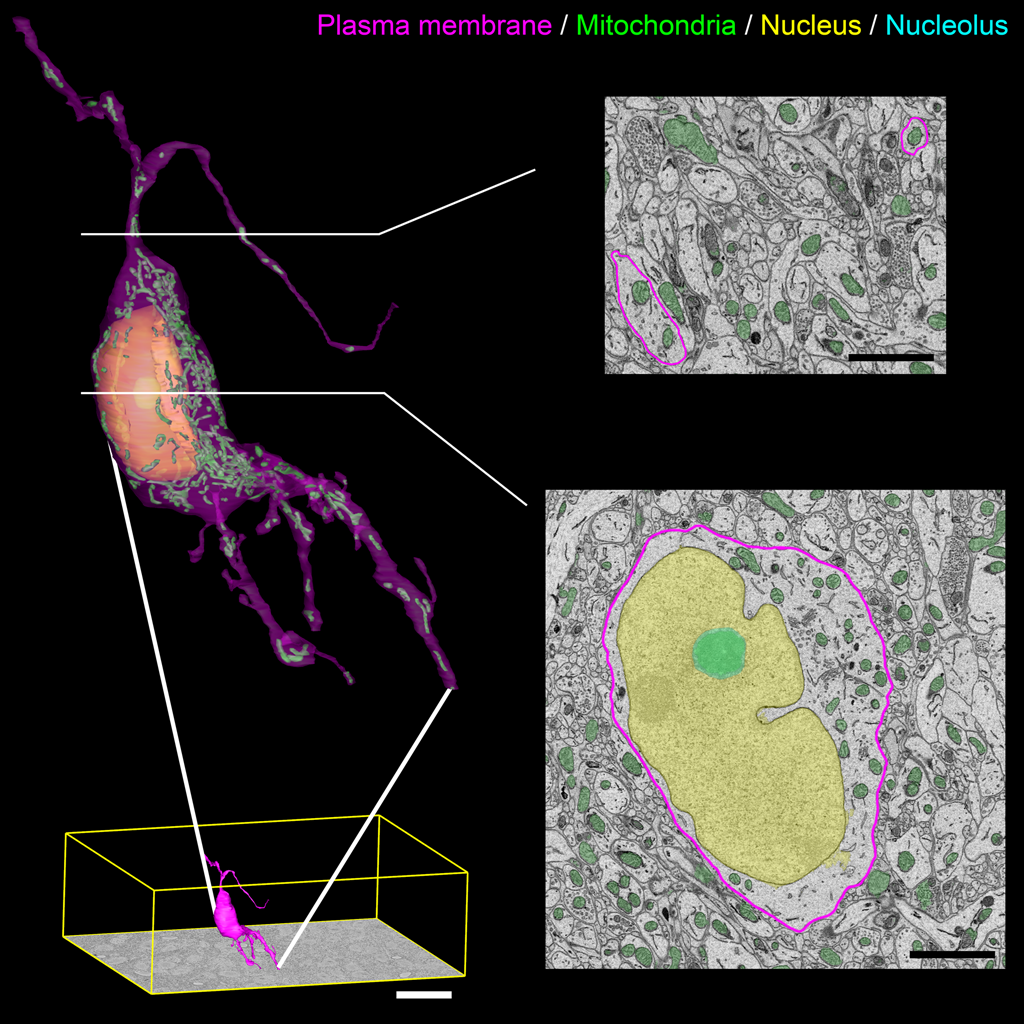
Figure Caption: Output surface renderings of manually segmented organelles within an SCN neuron. The plasma membrane of a neuron was manually traced in its entirety throughout the dataset. The size of this neuron with respect to the full dataset (bottom left, scale bar = 20 μm) demonstrates the scale of the segmentation challenge. An enlarged version of this neuron with a transparent plasma membrane is shown in the upper left corner. Surface renderings of the nucleus (yellow), nucleolus (cyan), and mitochondria (green) were generated from the output of our automatic segmentation workflow. Two cross-sectional planes through the neuron reveal the corresponding SBEM slice with transparent overlays of the probability maps for the three organelles (scale bar = 2 μm). Output renderings such as these can be used to analyze any number of parameters, including organelle morphology and clustering throughout the whole cell.
January 2015 La Jolla -- Electron microscopy (EM) is a very useful technique to analyze the form, distribution, and functional status of key organelle systems in various pathological processes including those associated with neurodegenerative diseases. Significantly, it has been used to provide important new insights into the mechanisms underlying diseases such as Parkinson’s and Alzheimer’s diseases and glaucoma.
Moreover, recent advances in EM instrumentation are fueling a renaissance in the study of quantitative neuroanatomy. Data obtained from techniques such as serial block-face scanning electron microscopy (SBEM) provide unprecedented volumetric snapshots of the in-situ biological organization of the mammalian brain across a multitude of scales. When combined with breakthroughs in specimen preparation, such data sets reveal not only a complete view of the membrane topography of cells and organelles, but also the location of cytoskeletal elements, synaptic vesicles, and certain macromolecular complexes. But while scientists are acquiring increasingly large volumes of 3D EM data, they are having trouble keeping up with the deluge of data: They need more efficient tools to measure precisely the 3D morphologies of organelles in data sets that can include hundreds to thousands of whole cells. Image segmentation and analysis remain significant bottlenecks to achieve quantitative descriptions of whole-cell structural composition.
Because manual segmentation is notoriously slow and labor-intensive, scientists’ ability to quantify and understand the details of subcellular components is dependent on advances in the accuracy, throughput, and robustness of automated segmentation methods. Computationally advanced methods for automated segmentation have produced increasingly accurate results and hold promise for still greater refinement. Moreover, while current computational methods yield impressive results, many are based on assumptions about the 3D morphology of the target organelle (e.g., mitochondria, nuclei, lysosomes). This is problematic not only because it makes extending such methods to segmentation of other organelles non-trivial but also because typical SBEM data sets contain a heterogeneous mixture of organelle morphologies across multiple cell types. So the bottom line is that scientists need a robust method to accurately segment various kinds of organelles in SBEM image stacks without any a priori assumptions about organelle morphology. In this context, a research team from UCSD, the University of Utah, and the Salk Institute for Biological Studies developed a novel method for the robust and accurate automatic segmentation of morphologically and functionally diverse organelles in EM image stacks.
This method trains organelle-specific pixel classifiers using the cascaded hierarchical model (CHM), a state-of-the-art, supervised, multi-resolution framework that uses only 2D image information. The CHM consists of bottom-up and top-down steps cascaded in multiple stages. Then a series of tunable 2D filters are applied to the outputs of the pixel classification to generate accurate segmentations. In the final processing step, connected components are meshed together in a manner that minimizes the deleterious effects of local and global imaging artifacts.
All training, pixel classification, and segmentation steps in this study were performed on a National Biomedical Computation Resource cluster, rocce.ucsd.edu (http://rocce-mgr.ucsd.edu/). All steps were performed using one CPU for each input image. One hundred CPUs were used, making it possible to expedite the work by processing 100 images in parallel. The team validated the method’s performance by assessing the accuracy of segmentation of four target organelles (mitochondria, lysosomes, nuclei, and nucleoli) in a sample SBEM data. These targets were chosen because they are morphologically and texturally diverse and, thus, pose a significant test of the robustness of the method. The robustness of the method was confirmed by demonstrating that the CHM performed very similarly when trained using either images from consecutive slices of the same nuclei or single slice images from a variety of nuclei. Moreover, the team parallelized their codes to segment all organelles in teravoxel-sized 3D EM data sets efficiently on supercomputing resources, resulting in a dramatic reduction in run time.
The resulting method, in summary, can be adapted easily to any organelle of interest. CHM classifiers can be trained for any data set or organelle target given the proper training data. And the method can be applied to teravoxel-sized data sets in a computationally efficient manner. The team estimates that a high-resolution, teravoxel-sized SBEM stack collected for an axon-tracking experiment can subsequently be downsampled and have its nuclei or mitochondria segmented automatically at a fraction of the computational cost that would have been required at native resolution. This reduction in computational cost will prove critical as further innovation leads to acquisition of ever larger data sets. The source code for CHM and all related scripts are available to download from http://www.sci.utah.edu/software/chm.html. Training images, training labels, and test images used in this study are also available at this URL.
This work was supported by grants from the National Institute of General Medical Science (NIGMS) under award P41 GM103412, the National Institute of Neurological Disorders and Stroke under award 1R01NS075314, the National Biomedical Computation Resource (NBCR) with support from NIGMS under award P41 GM103426, the National Institutes of Health (NIH) under award RO1 EY016807, and Fellowship support rom the National Institute on Drug Abuse under award 5T32DA007315-11.
Citation: Perez, Alex J., Mojtaba Seyedhosseini, Thomas J. Deerinck, Eric A. Bushong, Satchidananda Panda, Tolga Tasdizen, and Mark H. Ellisman, A workflow for the automatic segmentation of organelles in electron microcopy image stacks, Frontiers in Neuroanatomy (Methods Article), Vol. 8, Article 126, November 7, 2014, doi: 10.3389/fnana.2014.00126
Link to Article in PubMed Central