Research Highlights
CDeep3M—Plug-and-Play cloud-based deep learning for image segmentation
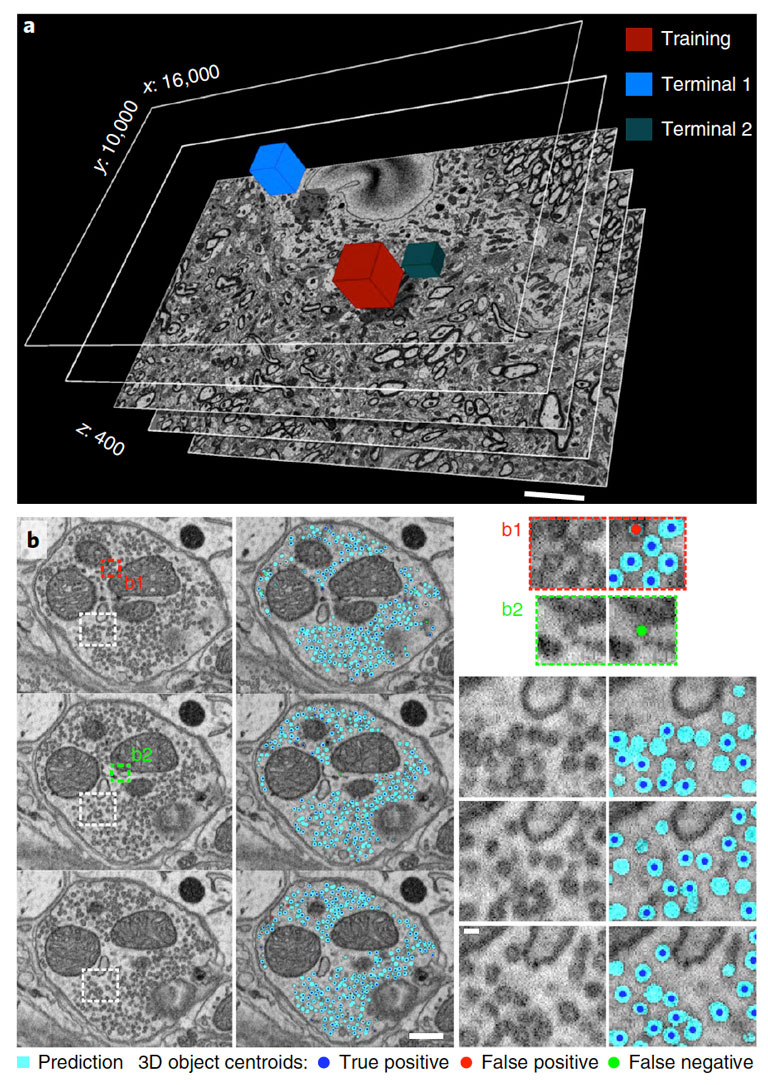
Synaptic vesicle counts on SBEM data using CDeep3M. a, SBEM volume acquired at 2.4 nm Å~ 24 nm voxel size (16,000 Å~ 10,000 Å~ 400 voxel). Performance tests were done on two terminals (Terminal 1: 3,184 vesicles and Terminal 2: 1,000 vesicles) comparing to several independent human counts. Scale bar, 5 μm. b, Three consecutive sections of Terminal 1 in overview (left panels, scale bar, 200 nm) alongside the CDeep3M predictions and zoomed in (right panels, scale bar, 40 nm) show comparisons to human counts. Centroids of 3D objects occurring across sections are marked on most prevalent plane.
31-08-2018 La Jolla
Deep neural networks, are a brain-inspired form of AI which are ideal for solving image analysis tasks. With rapid advances in biomedical imaging, users are now able to acquire images at ever increasing resolution and larger field of views. Manual analysis of such large datasets is no longer feasible and automated approaches are becoming critical to take full advantage of the increased data density. Deep learning, on the other hand, is quickly progressing and allowing new capabilities for identifying even the most challenging cellular features in these multi-terabyte datasets.
Computer algorithms can now automatically identify mitochondria, the power generators of a cell, or cell membranes. Unfortunately, the imaging community has struggled with making optimum use of these machine learning tools, especially at scale on large datasets. The complexity of tool deployment often requires researchers to be highly dependent on computer scientists to install, run and maintain complex software components on specialized high-end computer clusters. In their recent publication in Nature Methods, Haberl et al. (2018) addressed these bottlenecks by providing a ready-to-use image segmentation solution for any lab, with a publicly available deep convolutional neural network pre-configured for the Amazon cloud compute platform. “For us, the term ‘Open science’ moves beyond simply releasing code openly on GitHub. We want to make it easy for the community to deploy and use the code for their research and to reproduce our results” says Haberl “This is particularly important in the deep learning field which, in recent years, has increasingly struggled with reproducibility issues.”
These limitations are overcome with the CDeep3M cloud implementation, which gives end-users access to the same platform as the developers, along with the necessary software components installed and pre-configured. A simple ‘Launch’ button spins up the computer instance and installs the collection of required software for the end-user. By executing only three simple commands, users can train a deep neural network, and apply it to datasets in the terabyte range. The authors achieved high performance using CDeep3M on a variety of biomedical microscopy datasets, ranging from light, x-ray to cutting-edge electron microscopy acquisitions, with a resolution at the nm level using multi-tilt electron tomography and serial block-face scanning electron microscopy.
While cloud computing has become increasingly important in industrial applications, academia has been slow to deploy these technologies in production use. This is changing as the authors and others participate in new initiatives, like NIH’s Cloud Commons Credits pilot program, aimed to facilitate open access to specialized software products and to battle costs for maintenance of soft- and hardware. CDeep3M is the first of a series of specialized software products the researchers at NCMIR are making available as turn-key solutions through cloud computing platforms.
Funding Sources: This work was supported by NIH grants (5P41GM103412, 5P41GM103426 and 5R01GM082949) supporting the National Center for Microscopy and Imaging Research (NCMIR), the National Biomedical Computation Resource (NBCR), and the Cell Image Library (CIL). This research benefitted from the use of credits from the National Institutes of Health (NIH) Cloud Credits Model Pilot, a component of the NIH Big Data to Knowledge (BD2K) program. M.G.H. was supported by a postdoctoral fellowship from an interdisciplinary seed program at UCSD to build multiscale 3D maps of whole cells, called the Visible Molecular Cell Consortium. This work benefitted from the use of compute cycles on the Comet cluster, a resource of the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by National Science Foundation grant number ACI-1548562.
Relevant Publication: https://rdcu.be/5zIF
Haberl MG, Churas C, Tindall L, Boassa D, Phan S, Bushong EA, Madany M, Akay R, Deerinck TJ, Peltier ST, Ellisman MH. CDeep3M-Plug-and-Play cloud-based deep learning for image segmentation. Nat Methods. 2018 Sep;15(9):677-680. doi: 10.1038/s41592-018-0106-z. Epub 2018 Aug 31. PMID: 30171236
Additional Info: CDeep3M source code and documentation are available for download on GitHub https://github.com/CRBS/cdeep3m and is free for non-profit use.